Cloning Gates
Please note: this functionality requires FlowJo for Mac 9.3b5 or later.
Introduction
The use case for this feature came from our clinically inclined users who wanted one version of a gate propagated to different points in a hierarchy. This used to be done by one of two ways, neither of which is optimal in terms of workflow:
Create a version of the gate at a top level, and at each point in the tree, insert a boolean gate that refers solely to that top gate
Create the gate; copies of the gate are FJML where each vertex refers to the vertex of the original gate We implemented a new gate type, a "clone". Once you create a gate, you can choose to "clone" it, and apply it to different points
in the sample hierarchy. Both the original and all copies are then designated as a "clones". Whenever a clone is modified, all of its clones are also modified (including renaming it). Thus, a clone can be modified at any point in the tree, and all
of its versions will update appropriately.
Detailed Description
Any gate can be defined as a "clone". This is just a property of a gate. A clone can be unique in a sample, or there may be multiple clones of the gate in different places in the hierarchy. All clones in a sample will be identical; modify any one
of them and all update. To clone a gate, select it, and choose Workspace->Clone Gate. It can be un-cloned via the same menu operation.
A clone gate is designated by a special additional icon in front of the gate name (3 identical green circles) :  There can be no other gates with the same name as a "clone" gate in the sample. If you specify that "CD4" is a clone (in the /Lymphs/CD3 gate), then you can't have any other gate named "CD4" anywhere else in the hierarchy. Once you've defined "CD4"
as a clone, you can drag it to other places and clone it. (Alternatively, if you specify that "CD4" is a clone and there are other "CD4" gates, the program could allow you to force them to all become clones, I think that's reasonable). Note: If you drag a clone (e.g. "CD4") into a destination sample, then ALL "CD4" gates in that destination sample will automatically become clones.
There can be no other gates with the same name as a "clone" gate in the sample. If you specify that "CD4" is a clone (in the /Lymphs/CD3 gate), then you can't have any other gate named "CD4" anywhere else in the hierarchy. Once you've defined "CD4"
as a clone, you can drag it to other places and clone it. (Alternatively, if you specify that "CD4" is a clone and there are other "CD4" gates, the program could allow you to force them to all become clones, I think that's reasonable). Note: If you drag a clone (e.g. "CD4") into a destination sample, then ALL "CD4" gates in that destination sample will automatically become clones.
Brief Example
Let's link and display quad gates on all levels of a T cell analysis to show effect of different gates on contents of quads.
First, take a regular analysis with quads attached only in the relevant spot: 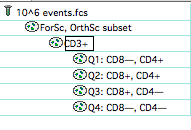 Select the quads and “clone” them from the Workspace menu:
Select the quads and “clone” them from the Workspace menu: 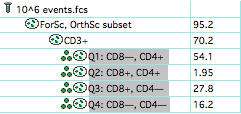 Next, copy the clones to other points in the hierarchy:
Next, copy the clones to other points in the hierarchy: 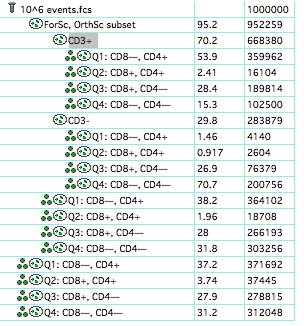 And finally display them in the layout:
And finally display them in the layout: 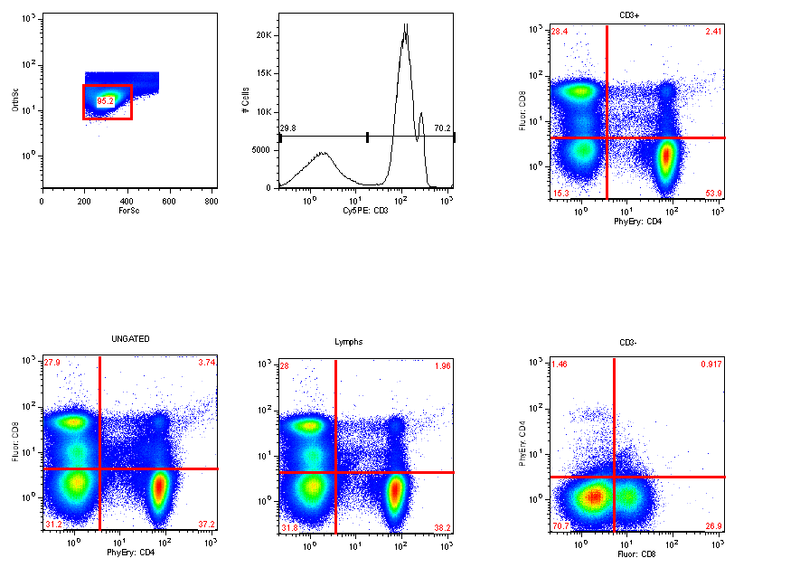 Now adjusting the quads in any graph window that displays this set, will update it for all 4 plots.
Now adjusting the quads in any graph window that displays this set, will update it for all 4 plots.





